Dinesh-Kumar Laboratory
I. Immune receptor function in pathogen recognition and activation of immune signaling
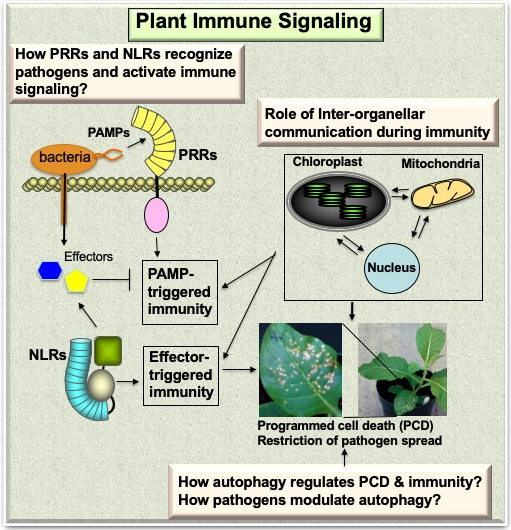
Similar to animals, plants rely on cell-surface and intracellular immune receptors to defend against pathogens. The front line of innate immunity in plants involves the recognition of highly conserved microbe/pathogen associated molecular patterns (MAMPs/PAMPs) such as bacterial flagellin or fungal chitin by cell-surface localized pattern recognition receptors (PRRs) leading to PAMP-triggered immunity (PTI) to limit pathogen growth. Highly evolved pathogens deliver effector proteins to interfere with the PTI response. Plants have evolved intracellular nucleotide-binding domain leucine-rich repeat (NLR) class of receptors which recognize pathogen effectors and activate effector-triggered immunity (ETI). The PTI and ETI pathways share many components; ETI has increased in the amplitude of the signals during ETI often culminates in hypersensitive response programmed cell death (HR-PCD) that physically isolates the infection. Our long-term goal is to understand the molecular mechanisms by which immune receptors recognize pathogens and initiate immune signaling. |
Pattern-triggered immunity: One of the main areas of research in our group is to understand how cell-surface PRRs recognize PAMPs to induce PTI. BIK1, a receptor-like cytoplasmic kinase (RLCK), functions as a central integrator of signaling from multiple PRRs. To understand how BIK1 decodes pathogen inputs into immune signaling outputs, we determined the BIK1 crystal structure. To integrate the BIK1 structure data, we mapped trans-phosphorylation sites of BIK1 by the PRR EFR, a receptor-like kinase that recognizes bacterial elongation factor peptide elf18 to activate immune signaling. Based on the trans-phosphorylation and structure data, we identified several important amino acid residues that could control immune signaling. Analyses of phosophonull and phosphomimetic transgenic plants identified S89 and T90 residues in BIK1 play important roles in response to Pseudomonas bacterial infection. Interestingly, the S89 and T90 reside in a uniquely extended loop in the BIK1 structure compared to homologous kinases. Phosphonull mutants were more susceptible and phosphomimetic mutants were more resistant to bacterial infection. Interestingly, S89D/T90d phosphomimetic mutants have increased levels of the phytohormone jasmonic acid (JA), which plays a role in immune signaling. BIK1 also localizes to the nucleus where it phosphorylates WRKY transcription factors to regulate JA and SA hormone responsive genes. These findings provided the mechanistic basis of signal transduction from PRR to phytohormones during immune signaling (PMC6266874).
Extracellular reactive oxygen species (ROS) produced through NADPH oxidase RbohD plays an important role in PRR-mediated immune signaling to limit pathogen infection. We recently contributed towards understanding the role of MAPKKKK (MAP4K) called SIK1 that interacts and stabilizes BIK1 during PRR signaling and promotes ROS burst through RbohD (PMC6279242). In another collaborative work with Professor Gitta Coaker laboratory, we demonstrated that RbohD activity is switched off after initiation of PRR signaling through phosphorylation of RbohD by PBL13 kinase and RbohD ubiquitination through a previously uncharacterized E3 ubiquitin ligase PIRE1 (PMC7160206).
Effector-triggered immunity: Another main area of research in our group is to understand how intracellular NLRs recognize pathogen effectors and activate defense signaling. To this end, we study N immune receptor-mediated defense against Tobacco Mosaic Virus (TMV) as a model. N is the first member of plant Toll-Interleukin-1 receptor homology Region (TIR) domain containing NLR class of immune receptor (TIR-NLR). N is a nucleocytoplasmic receptor and recognizes the helicase domain (referred to as p50) within the TMV replicase. N recognizes p50 indirectly through a chloroplast localized N Receptor Interacting Protein 1 (NRIP1). In the presence of TMV, a subset of available NRIP1 relocalizes from the chloroplast to the cytoplasm and nucleus. Cytoplasmic NRIP1 associates with the TMV replicase leading to the recruitment of N (PMC1820829; PMC2267721). Interestingly, N associates directly with a transcription factor, Squamosa Promoter-binding-protein-Like 6 (SPL6), and this association occurs only upon recognition of the p50 effector. Silencing of SPL6 compromises N-mediated resistance to TMV. SPL6 is also required for Arabidopsis TIR-NLR RPS4-mediated resistance to Pseudomonas syringae bacteria expressing the AvrRps4 effector. These findings point to SPL6 as one of the conserved nuclear components of TIR-NLR signaling (PMC3597514).
Recently, we optimized and used a proximity-labeling technique in plants and identified number of novel proteins which play a role in N-mediated resistance to TMV. Our findings indicated that a novel UBR-box containing E3 ubiquitin ligase, UBR7, plays an important role in modulating N protein levels and immune response to TMV (PMC6642208).
Extracellular reactive oxygen species (ROS) produced through NADPH oxidase RbohD plays an important role in PRR-mediated immune signaling to limit pathogen infection. We recently contributed towards understanding the role of MAPKKKK (MAP4K) called SIK1 that interacts and stabilizes BIK1 during PRR signaling and promotes ROS burst through RbohD (PMC6279242). In another collaborative work with Professor Gitta Coaker laboratory, we demonstrated that RbohD activity is switched off after initiation of PRR signaling through phosphorylation of RbohD by PBL13 kinase and RbohD ubiquitination through a previously uncharacterized E3 ubiquitin ligase PIRE1 (PMC7160206).
Effector-triggered immunity: Another main area of research in our group is to understand how intracellular NLRs recognize pathogen effectors and activate defense signaling. To this end, we study N immune receptor-mediated defense against Tobacco Mosaic Virus (TMV) as a model. N is the first member of plant Toll-Interleukin-1 receptor homology Region (TIR) domain containing NLR class of immune receptor (TIR-NLR). N is a nucleocytoplasmic receptor and recognizes the helicase domain (referred to as p50) within the TMV replicase. N recognizes p50 indirectly through a chloroplast localized N Receptor Interacting Protein 1 (NRIP1). In the presence of TMV, a subset of available NRIP1 relocalizes from the chloroplast to the cytoplasm and nucleus. Cytoplasmic NRIP1 associates with the TMV replicase leading to the recruitment of N (PMC1820829; PMC2267721). Interestingly, N associates directly with a transcription factor, Squamosa Promoter-binding-protein-Like 6 (SPL6), and this association occurs only upon recognition of the p50 effector. Silencing of SPL6 compromises N-mediated resistance to TMV. SPL6 is also required for Arabidopsis TIR-NLR RPS4-mediated resistance to Pseudomonas syringae bacteria expressing the AvrRps4 effector. These findings point to SPL6 as one of the conserved nuclear components of TIR-NLR signaling (PMC3597514).
Recently, we optimized and used a proximity-labeling technique in plants and identified number of novel proteins which play a role in N-mediated resistance to TMV. Our findings indicated that a novel UBR-box containing E3 ubiquitin ligase, UBR7, plays an important role in modulating N protein levels and immune response to TMV (PMC6642208).
II. Role of inter-organellar communications during innate immunity
Another active area of research in the lab is to understand the role of inter-organellar communication during immunity. As described above, chloroplast localized NRIP1 is recruited to the cytoplasm and nucleus to mediate pathogen recognition and activation of immune signaling. Interestingly, during the immune response chloroplasts send out dynamic tubular projections known as “stromules”. This suggests that chloroplasts have an additional function downstream of pathogen recognition. Stromules are induced during the beginning phases of PCD and pro-PCD cell-to-cell signals such as hydrogen peroxide (H2O2) and salicylic acid (SA) are sufficient to induce stromule formation. During the immune response, stromules form strong connections with nuclei. These connections precede the accumulation of chloroplast-localized NRIP1 and H2O2 in the nucleus. These data provide evidence that stromule to nuclei connections are involved in the signaling from chloroplasts to nuclei during a defense response. Furthermore, we showed that constitutive induction of stromules enhances PCD, proving that stromules function during the progression of PCD. We propose a model in which stromules are involved in transducing and amplifying pro-defense signals originating from chloroplasts (PMC4596411).
In a recent paper, we show that chloroplast stromules dynamically extend along microtubules and anchor to a fine network of actin microfilaments. Stromules appear to guide the movement of chloroplasts, and our work uncovered this hitherto unknown role of stromules and defined a newly discovered form of chloroplast movement. We show that stromules are anchored to actin filaments along nuclei, and these interactions guide the clustering of chloroplasts to nuclei during plant innate immunity (PMC5815851). Future studies will determine if stromules just guide chloroplast movement or provide a physical force that drives the movement.
Another active area of research in the lab is to understand the role of inter-organellar communication during immunity. As described above, chloroplast localized NRIP1 is recruited to the cytoplasm and nucleus to mediate pathogen recognition and activation of immune signaling. Interestingly, during the immune response chloroplasts send out dynamic tubular projections known as “stromules”. This suggests that chloroplasts have an additional function downstream of pathogen recognition. Stromules are induced during the beginning phases of PCD and pro-PCD cell-to-cell signals such as hydrogen peroxide (H2O2) and salicylic acid (SA) are sufficient to induce stromule formation. During the immune response, stromules form strong connections with nuclei. These connections precede the accumulation of chloroplast-localized NRIP1 and H2O2 in the nucleus. These data provide evidence that stromule to nuclei connections are involved in the signaling from chloroplasts to nuclei during a defense response. Furthermore, we showed that constitutive induction of stromules enhances PCD, proving that stromules function during the progression of PCD. We propose a model in which stromules are involved in transducing and amplifying pro-defense signals originating from chloroplasts (PMC4596411).
In a recent paper, we show that chloroplast stromules dynamically extend along microtubules and anchor to a fine network of actin microfilaments. Stromules appear to guide the movement of chloroplasts, and our work uncovered this hitherto unknown role of stromules and defined a newly discovered form of chloroplast movement. We show that stromules are anchored to actin filaments along nuclei, and these interactions guide the clustering of chloroplasts to nuclei during plant innate immunity (PMC5815851). Future studies will determine if stromules just guide chloroplast movement or provide a physical force that drives the movement.
III. Role of autophagy in programmed cell death (PCD), immunity and pathogenesis
Another active area of research in our group is to understand the regulation of PCD induced during the immune response. We discovered that macroautophagy (hereinafter referred to as autophagy) plays an important role in delimiting the PCD to the infection site (PMID:15907470; PMID:17932459). Autophagy is a dynamic process conserved across eukaryotes that entails the engulfment of cellular components in double membrane vesicles called autophagosomes that are then targeted to the vacuole/lysosome for degradation or recycling. Autophagosome formation and cargo delivery is regulated by a series of Autophagy (Atg) proteins that are conserved from yeast to higher eukaryotes including plants. To understand the interplay between autophagy and PCD during immunity, we have been studying Atg-interacting proteins and different cargoes carried by autophagosomes during immune response. Atg8 plays an important role in autophagosome biogenesis and recruitment of cargo to autophagosomes. Biogenesis of autophagosomes requires processing of Atg8 by Atg4, a cysteine protease, and conjugation of PE (phosphatidylethanolamine) to Atg8. While yeast has only one Atg4 and one Atg8, the Arabidopsis genome contains two Atg4 (AtAtg4a and AtAtg4b) and nine Atg8 (AtAtg8a–AtAtg8i) homologs.
Using a novel BRET-based synthetic AtAtg8 substrate combined with biochemical studies, we showed that AtAtg4a is predominantly involved in processing and production of mature AtAtg8 isoforms (PMC3896200). This hints at a potential selective function for the different AtAtg8 isoforms in planta during different biological processes. Our recent sequence and phylogenetic analyses from 17 sequenced plant genomes indicate that Atg4 and Atg8 proteins are highly conserved in plant lineages. Our analyses suggested that the Atg8 expansion in plants might be attributed to whole genome duplication, segmental and dispersed duplication, and purifying selection. Our cross-kingdom biochemical analyses of Atg8 processing by Atg4 indicated that human Atg4 has broad substrate specificity and cleaves Atg8s from yeast, plant and human; whereas, plant and yeast Atg4s fails to process human Atg8 (also known as LC3a). Molecular modeling indicated that the lack of processing of HsLC3A is due to structural differences between HsLC3A and yeast and plant Atg8s (PMC5103345).
Another active area of research in our group is to understand the regulation of PCD induced during the immune response. We discovered that macroautophagy (hereinafter referred to as autophagy) plays an important role in delimiting the PCD to the infection site (PMID:15907470; PMID:17932459). Autophagy is a dynamic process conserved across eukaryotes that entails the engulfment of cellular components in double membrane vesicles called autophagosomes that are then targeted to the vacuole/lysosome for degradation or recycling. Autophagosome formation and cargo delivery is regulated by a series of Autophagy (Atg) proteins that are conserved from yeast to higher eukaryotes including plants. To understand the interplay between autophagy and PCD during immunity, we have been studying Atg-interacting proteins and different cargoes carried by autophagosomes during immune response. Atg8 plays an important role in autophagosome biogenesis and recruitment of cargo to autophagosomes. Biogenesis of autophagosomes requires processing of Atg8 by Atg4, a cysteine protease, and conjugation of PE (phosphatidylethanolamine) to Atg8. While yeast has only one Atg4 and one Atg8, the Arabidopsis genome contains two Atg4 (AtAtg4a and AtAtg4b) and nine Atg8 (AtAtg8a–AtAtg8i) homologs.
Using a novel BRET-based synthetic AtAtg8 substrate combined with biochemical studies, we showed that AtAtg4a is predominantly involved in processing and production of mature AtAtg8 isoforms (PMC3896200). This hints at a potential selective function for the different AtAtg8 isoforms in planta during different biological processes. Our recent sequence and phylogenetic analyses from 17 sequenced plant genomes indicate that Atg4 and Atg8 proteins are highly conserved in plant lineages. Our analyses suggested that the Atg8 expansion in plants might be attributed to whole genome duplication, segmental and dispersed duplication, and purifying selection. Our cross-kingdom biochemical analyses of Atg8 processing by Atg4 indicated that human Atg4 has broad substrate specificity and cleaves Atg8s from yeast, plant and human; whereas, plant and yeast Atg4s fails to process human Atg8 (also known as LC3a). Molecular modeling indicated that the lack of processing of HsLC3A is due to structural differences between HsLC3A and yeast and plant Atg8s (PMC5103345).
IV. Development of virus-induced gene silencing (VIGS) and CRISPR/Cas9-mediated genome editing approaches for gene function studies
VIGS is an RNA-silencing based technique used for the targeted downregulation of a host gene, allowing the analysis of the gene’s function. VIGS is rapid (1-2 weeks from infection to silencing), does not require the development of stable transformants, and is amenable to high throughput forward and reverse genetic screening. We have developed and used an improved Tobacco rattle virus (TRV)-based VIGS system (PMID:12028572; PMID:12220268; PMID:15356389; PMC1557620; PMC2151726; PMC6508541) to study gene function in plants. The TRV system is the most widely used viral vector in plants and its utility has been demonstrated in over 25 plant species including a large number of plants in Solanaceae, as well as in Brassicaceae, Euphorbiaceae, Malvaceae, Papaveraceae and Rosaceae..
The CRISPR/Cas9 system has been used for efficient targeted genome editing in various organisms including plants. To date, most published reports in plants require the generation of transgenic plants to deliver Cas9 and sgRNA. Transgenic plant production is time consuming and some crop plants are recalcitrant to transformation. Therefore, efficient delivery of Cas9 and sgRNA into plant cells is required for rapid crop trait discovery that circumvents the requirement of transformation. Therefore, one of the active research areas in our group is to engineer viral vectors to deliver Cas9 and sgRNA to induce genome editing. We have recently demonstrated in a collaborative project that our TRV-based system could be used to deliver sgRNA into Cas9 expressing plants to induce heritable genome editing at a low frequency (PMID:25749112). Currently, we are modifying TRV vectors and optimizing methods to improve the efficiency of heritable genome editing and to facilitate the generation of non-transgenic genome modification in plants.
VIGS is an RNA-silencing based technique used for the targeted downregulation of a host gene, allowing the analysis of the gene’s function. VIGS is rapid (1-2 weeks from infection to silencing), does not require the development of stable transformants, and is amenable to high throughput forward and reverse genetic screening. We have developed and used an improved Tobacco rattle virus (TRV)-based VIGS system (PMID:12028572; PMID:12220268; PMID:15356389; PMC1557620; PMC2151726; PMC6508541) to study gene function in plants. The TRV system is the most widely used viral vector in plants and its utility has been demonstrated in over 25 plant species including a large number of plants in Solanaceae, as well as in Brassicaceae, Euphorbiaceae, Malvaceae, Papaveraceae and Rosaceae..
The CRISPR/Cas9 system has been used for efficient targeted genome editing in various organisms including plants. To date, most published reports in plants require the generation of transgenic plants to deliver Cas9 and sgRNA. Transgenic plant production is time consuming and some crop plants are recalcitrant to transformation. Therefore, efficient delivery of Cas9 and sgRNA into plant cells is required for rapid crop trait discovery that circumvents the requirement of transformation. Therefore, one of the active research areas in our group is to engineer viral vectors to deliver Cas9 and sgRNA to induce genome editing. We have recently demonstrated in a collaborative project that our TRV-based system could be used to deliver sgRNA into Cas9 expressing plants to induce heritable genome editing at a low frequency (PMID:25749112). Currently, we are modifying TRV vectors and optimizing methods to improve the efficiency of heritable genome editing and to facilitate the generation of non-transgenic genome modification in plants.