Current Projects
Zygotic Genomic Activation in Rice
PI: Venkatesan Sundaresan, University of California, Davis
Co-PI: Scott Russell and his Grad. Student, Daniel Jones, Department of Molecular and Plant Biology, University of Oklahoma, Norman, Oklahoma
The fusion of two highly differentiated cells, the egg cell and the sperm cell, results in a totipotent zygote: a single cell with the capacity to develop into an entirely new organism. In this respect, the zygote can be considered to be the ultimate stem cell. In both plants and animals, early embryo development is dependent on maternal transcripts deposited in the egg cell before fertilization. The degradation of these maternal transcripts and the initiation of zygotic transcription take place during the Maternal to Zygotic Transition (MZT). Regulation of the MZT in plants and animals is critical to successful reproduction.
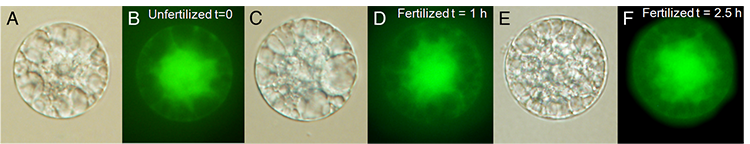
Isolated Rice Zygotes
We are studying the transcriptomes of rice zygotes to answer the following questions: When and how does the maternal to zygotic transition occur in rice.
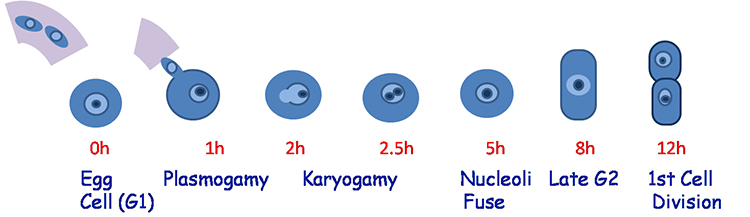
Time Course: Fertilization and the 1st Cell Division of the Rice Zygote
We are studying the transcriptomes of rice zygotes to answer the following questions: When and how does the maternal to zygotic transition occur in rice, and what effect do parental contributions play in this process?

Rice has several characteristics that make it suitable for this project. First, rice has the shortest time from pollination to fertilization of the model plants, just 30 minutes vs. several hours in Arabidopsis or maize. This allows for the precise determination of developmental time points. Second, relatively larger size, living rice zygotes can be dissected, making analysis of specific time points possible. Moreover, the availability of genomic resources including high-quality annotation in rice facilitates large-scale genomic analysis. We are characterizing the changes in the transcriptome starting with the very first cell cycle of the plant embryo, and plan to identify the individual contributions of the maternal and paternal genomes to the zygotic transcriptome. This project should result in a first detailed analysis of a critical transition in the plant life cycle, and reveal some of the parental and zygotic factors that control the process.
Go to full project page
Patterning the Female Gametophyte in Arabidopsis
Development of the gametophyte, the haploid generation of the plant life-cycle, is important for plant reproduction and seed formation. In flowering plants, the female gametophyte is called the embryo sac. It consists of only four cell types, including two gametes, which are the result of a precise developmental program.
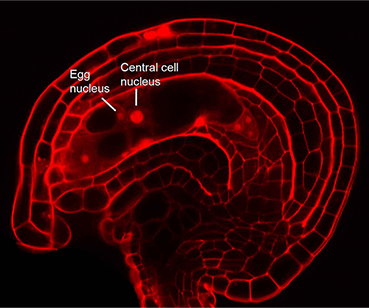
How flowering plants specify their female gametes: Patterning of the Arabidopsis Embryo Sac
Until recently, relatively little was known about the molecular mechanisms that establish developmental pattern in the embryo sac, in part due to the relative inaccessibility of the gametophyte and the limited availability of informative mutants. Experimental evidence has now been obtained that signaling by the hormone auxin, acting through an asymmetric distribution in the ovule, is the key to position based cell identity in the embryo sac.
The effects of auxin on cell specification in the embryo sac are being investigated through genetic and molecular manipulations, as well as interactions with other hormonal pathways. These studies will provide an understanding of the mechanisms underlying pattern formation, and how the two gametes, the egg cell and the central cell are specified. Hydroponically grown rice will be analyzed for changes in gene expression. Because of the key role of the embryo sac in reproduction, a deeper understanding of the female gametophyte can yield potential applications towards the control of reproduction and seed production in crop plants.
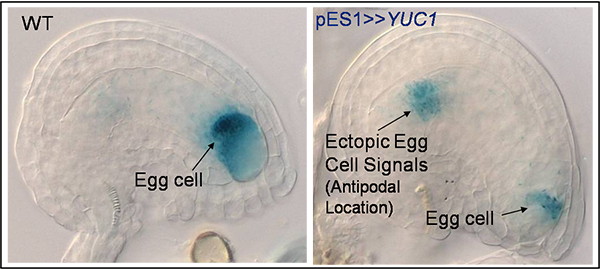
Upregulating auxin levels results in ectopic egg cell fate at opposite poles of the Arabidopsis Embryo Sac.
Result: Ectopic egg cell at opposite pole of Embryo Sac
Genomics of Host-Microbiome Interactions in Rice
PI: Venkatesan Sundaresan, University of California, Davis
Co-PI: Jonathan Eisen, Genome Center, University of California, Davis
Co-PI: Davis Mackill, Department of Plant Sciences, University of California, Davis
Co-PI: Merle Anders, Crop, Soil and Environmental Sciences, University of Arkansas, Fayetteville
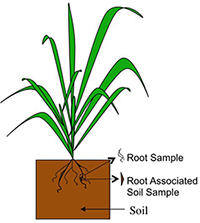
Schematic showing the rhizosphere around a rice plant's roots
Background
Plants, much like humans and other animals, harbor rich communities of potentially beneficial bacteria. The collective genomes of this complex microbial network, termed the metagenome, encode diverse metabolic capabilities unfounded within plants, essentially offering a functional extension to the host plant's genome.

A soil sample from field grown rice plants for rhizosphere analysis
Soil microbes in close proximity to plant roots (termed the rhizosphere) have been shown to promote plant disease suppression and nutrient acquisition.
Using rice (Oryza sativa) as a model, we are answering questions based around how plants recruit and moderate their associated root microbiomes.Unraveling the composition and maintenance of the rice microbiome is not only important for bolstering our basic understanding of host-microbe interactions: it is of great agronomic and ecological importance as well. Rice, due to its world-wide cultivation and consumption, is recognized as one of the most important agriculturally grown cereals. Understanding how broad communities of soil bacteria impact rice plant performance and yield will provide new directions for agricultural technologies.
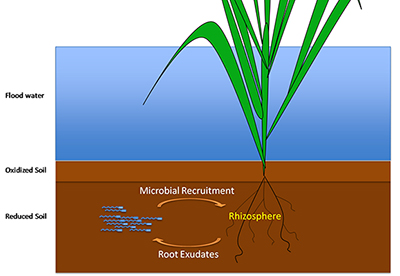
Recruitment of microbial communities by root exudates from rice roots in a paddy field
Rice cultivation also has ecological impacts. Rice paddies account for 15 – 20% of global methane emissions. Methane (a greenhouse gas) is produced by methanogenic archaea that feed on organic material in the paddy soil, including root exudates and decaying root material. The produced methane is taken up by the rice plant and emitted through a specialized gas vascular system known as aerenchyma. Interestingly, the rice plant inherently provides an environment conducive for methane-utilizing bacteria in the rhizosphere by partially oxygenating the soil adjacent to the roots, providing a substrate for methane oxidation.
Understanding mechanisms for how a rice plant moderates levels of these methane-utilizing bacteria is of critical environmental importance.
Go to full project page
Past Projects
The Rice Microbiome and its effect on the Transcriptome

Schematic showing the rhizosphere around a rice plant's roots
Background
Plants, much like humans and other animals, harbor rich communities of potentially beneficial bacteria. The collective genomes of this complex microbial network, termed the metagenome, encode diverse metabolic capabilities unfounded within plants, essentially offering a functional extension to the host plant's genome.

A soil sample from field grown rice plants for rhizosphere analysis
Soil microbes in close proximity to plant roots (termed the rhizosphere) have been shown to promote plant disease suppression and nutrient acquisition.
Using rice (Oryza sativa) as a model, we are answering questions based around how plants recruit and moderate their associated root microbiomes.Unraveling the composition and maintenance of the rice microbiome is not only important for bolstering our basic understanding of host-microbe interactions: it is of great agronomic and ecological importance as well. Rice, due to its world-wide cultivation and consumption, is recognized as one of the most important agriculturally grown cereals. Understanding how broad communities of soil bacteria impact rice plant performance and yield will provide new directions for agricultural technologies.

Recruitment of microbial communities by root exudates from rice roots in a paddy field
Rice cultivation also has ecological impacts. Rice paddies account for 15 – 20% of global methane emissions. Methane (a greenhouse gas) is produced by methanogenic archaea that feed on organic material in the paddy soil, including root exudates and decaying root material. The produced methane is taken up by the rice plant and emitted through a specialized gas vascular system known as aerenchyma. Interestingly, the rice plant inherently provides an environment conducive for methane-utilizing bacteria in the rhizosphere by partially oxygenating the soil adjacent to the roots, providing a substrate for methane oxidation.
Understanding mechanisms for how a rice plant moderates levels of these methane-utilizing bacteria is of critical environmental importance.
Specific Projects and Aims
i) Rice Plant Transcriptional Response to Microbiome
Recent advances in metagenomic methodologies and sequencing technologies have allowed us to start answering questions involving how a plant transcriptionally responds to or recruits a microbiome.
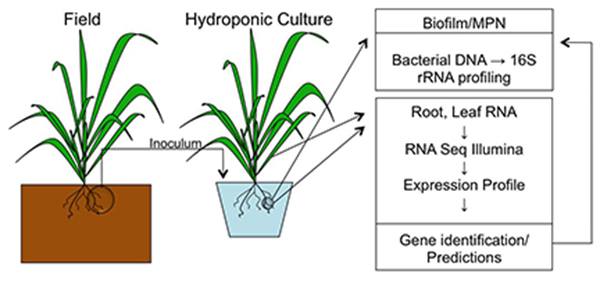
Hydroponically grown rice will be analyzed for changes in gene expression.
ii) Composition and Fluctuation of Rice Root Microbiome
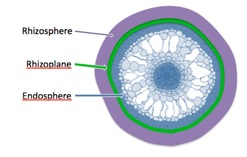
Root Compartments
Factors that influence the composition of rice root-associated microbiomes
Our deep-sequencing studies under controlled greenhouse conditions as well as in field conditions, using rice fields from several locations in Northern California have provided us with a wealth of microbial sequence data. Using these data, we have determined the key factors that cause variations in the taxonomic composition of the microbiome:
- Soil source, which provides the pool of microbes from which the plant recruits its microbiome. But even though the individual taxa might vary, the phyla that inhabit the different compartments of the microbiome do not change much with different soils.
- Genotype of the plant results in variation in the microbiome composition. Indica and japonica cultivars (the two most widely cultivated rice varieties) can be separated based on their microbiomes. This means that genetic factors influence the taxa that are recruited by the plant, an effect that can be exploited for plant breeding to recruit beneficial microbes.
- Geographical variation, which is likely to be based on climatic factors, can be observed in the microbiomes of rice plants collected from different fields in Northern California.
- Cultivation practice affects the microbiomes. We collaborated with Lundberg Farms to compare samples from organic and conventionally cultivated rice fields from different locations in Northern California. We find compositional differences between the two types of cultivation, reflected even in the occurrence of certain phyla representing potentially beneficial bacteria.
- Plant stress. We have found very rapid changes in the root microbiomes in response to both biotic and abiotic stresses. This suggests that microbiomes might have a role in the adaptive responses of plants to external and internal stresses.
Microbial Consortia
The very large datasets we have assembled can be exploited to reveal groups of microbes that might form "consortia", i.e., groups of microbes that form biological associations. For example, we have been able to identify potential consortia in methane cycling, that consist of methane producing archaea (methanogens) and their supporting eubacteria, as well as methane-consuming eubacteria (methanotrophs). This is a promising approach to identify bacterial communities that promote specific biological processes or pathways that are potentially beneficial to the plant.
Plant Response to Microbiomes
An axonic flask system has been established to grow rice plants in the presence and absence of a standardized stable microbe population. The response of the rice plants to these microbes is being assessed by transcriptomics using next generation sequencing techniques.
The specific genes determined to be up or down regulated in the presence of the microbes will be used to infer plant microbe crosstalk and help establish an understanding of the underlying symbiotic networks.
Relevant publications
- Spence, C., Alff, E., Johnson, C., Ramos, C., Donofrio, N., Sundaresan,V., Bais, H., 2014. Natural rice rhizospheric microbes suppress rice blast infections. BMC Plant Biology, 14:130.
- Yang S-Y, Gronlund M, Jakobsen I, Grotemeyer MS, Rentsch D, Miyao A, Hirochika H, Kumar CS, Sundaresan V, Salamin N, Catausan S, Mattes N, Heuer S, Paszkowski U. 2013. Nonredundant Regulation of Rice Arbuscular Mycorrhizal Symbiosis by Two Members of the PHOSPHATE TRANSPORTER1 Gene Family. The Plant Cell, 24: 4236-4251.
- Gutjahr C, Radovanovic D, Geoffroy J, Zhang Q, Siegler H, Chiapello M, Casieri L, An K, An G, Guiderdoni E, Kumar CS, Sundaresan V, Harrison MJ, Paszkowski U. 2011. The half-size ABC transporters STR1 and STR2 are indispensable for mycorrhizal arbuscule formation in rice. The Plant Journal 69: 906-920.
Functional Genomics of Model Plants
Introduction
We have been using insertional mutagenesis for the identification of gene expression pattern and function in plants.
Our approach utilizes gene and enhancer trap mutagenesis by transposons (Sundaresan et al. 1995), which together with
sequencing of flanking DNA to establish an FST database (Parinov et al. 1999), provides a powerful tool for functional
genomics (Parinov and Sundaresan 2000; Ramachandran and Sundaresan 2000). The original application of this strategy was
in Arabidopsis. We are now applying this strategy in rice, which is an important model plant for monocots, and whose
genome has been recently sequenced. A high-efficiency strategy for generating large numbers of transposon knockouts
in rice has been established in our laboratory (Kolesnik et al. 2004; Kumar et al. 2005), which will be useful for the
large scale analysis of gene function in rice as well as other cereal crops. A second area of research in functional
genomics is the identification of small RNAs in rice and maize. This is a component of the transcriptome which has an
important function in the control of gene expression as well as in epigenetic silencing, whose importance has only been
recognized in the past few years. We have developed computational methods for identification of micro RNAs in Arabidopsis
(Adai et al. 2005). Now we are extending these methods to rice and maize, in collaboration with Dr. Vicki Vance and Dr.
Lew Bowman (U. South Carolina) who are generating large numbers of small RNA sequences from these plants. These studies
will eventually lead to a map of the small RNA transcriptome in large plant genomes to identify their specific and global functions.
Abstract
Rice has a relatively small genome of about 430 Mb, which is the second plant genome to be completely sequenced. Rice is also
amenable to genetic studies and transformation, and is not only a model system for genome studies of the cereals but also an
important crop in its own right. Currently there are very limited tools for the identification of functions of sequenced genes
in rice that are widely available to the research community. In this project, we will develop efficient transposon tagging strategies
for large scale transposon mutagenesis in rice. Stable insertion lines will be generated. Containing random insertions of the maize
En/Spm or Ac-Ds transposons in japonica rice cv. Nipponbare, which is the cultivar selected for the genome sequence. A public database
for reverse genetics containing sequences of the DNA flanking the insertions will be established, and the sequences will be linked to
the rice genome database and the rice genome physical map. This effort should lead to a significant resource for functional genomics
in rice, as well as other cereals. The project also provides a training component for undergraduate students from the three
participating universities.
Goals
- To establish high efficiency insertional mutagenesis for rice using an engineered transposon system.
- To generate a population of tagged lines containing independent transposon insertions.
- To determine the flanking sequences and physical locations for 5,000+ of the transposon insertions, and to generate a
searchable database of insertion sites.
Availability of materials and information
The flanking sequence database will be publicly accessible through a website to be maintained by the Arizona Genomics
Institute, which will be linked to the rice genome databases. All of the sequenced insertion lines will be released to
the research community by the end of the 3 year funding period.
Training and Diversity
The project is providing training for postdoctoral fellows and several undergraduate students.
There is a program for training visiting undergraduate students from Grand Valley State University,
Michigan selected by Dr. Regina McClinton, and from University of Lousiana-Lafayette, selected by Dr. Caryl Chlan.
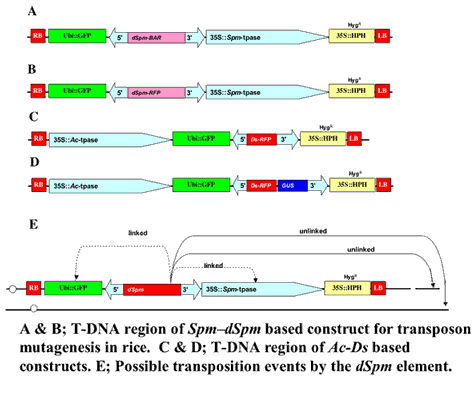
Two component transposon tagging systems for rice
- System is based on maize Suppressor mutator (Spm) transposon
35S :: Spm-transposase
dSpm :: RFP
- System is based on maize Activator (Ac) transposon
35S :: Ac-transposase
Ds :: RFP or Ds Gene trap :: (RFP; GUS)
GFP as negative selection marker, RFP as positive selection marker & HPH as marker for selecting transformants

T-2 Plants in the field, 2004
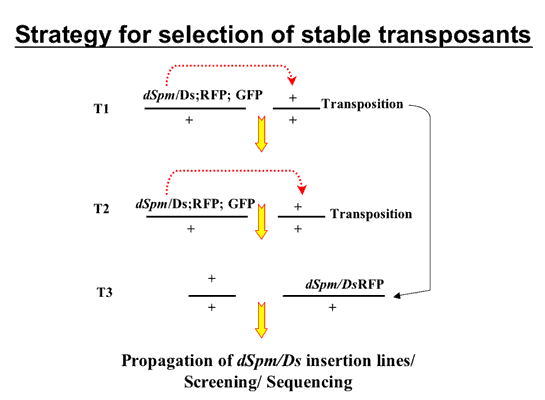
Selection of stable transposants using the positive and negative selection system
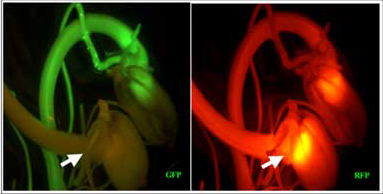
StableT3 transposants are selected from the progeny of Heterozygous T2 plants based on the absence of GFP, and the presence of RFP fluoresence

High throughput transposon screening using seedlings in 96 well plates and Kodak Image Station 2000MM. Arrows indicate a transposant which is GFP- (left) and RFP+ (right)
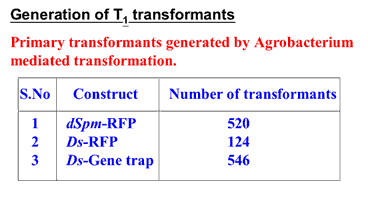
Rice transposon insertion library (As of September 15, 2008)
| No |
Line ID |
No. of lines Generated |
No. of lines Sequenced |
T4 Seeds for Distribution |
Submitted to Genbank |
| 1 |
RGT |
7803 |
5905 |
3037 |
3612 |
| 2 |
RDs |
2014 |
1879 |
899 |
1124 |
| 3 |
RdSpm |
20277 |
13645 |
5865 |
13094 |
| 4 |
ADs |
970 |
559 |
- |
- |
| Total |
31064 |
21988 |
9801 |
17940 |
Related links
Genetics of Gametogenesis in Arabidopsis
Introduction
Mutant and expression screens have provided a rich harvest of information on genes regulating fundamental plant processes. They include several genes regulating vegetative and reproductive development characterized by our laboratory (e.g. Yang et al. 1999; Tantikanjana et al. 2000; Rajani and Sundaresan, 2001; Kumaran et al. 2002). The current focus of our laboratory is on genes that are required for gametogenesis and early embryogenesis, many of which are genes involved in the regulation of cell cycle or cell division patterns (e.g. Yang and Sundaresan, 2000). In collaboration with Dr. Sheila McCormick (USDA) we have characterized gametophyte mutants in Arabidopsis that were generated in our laboratory, leading to the identification of over 100 genes involved in various aspects of embryo sac development and function (Pagnussat et al. 2005). A complementary strategy using microarrays has also been performed to identify another set of over 200 genes with potential functions in the female gametophyte (Yu et al. 2005). These studies help us understand a critical step in plant reproduction, and potentially lead to applications in agriculture through control of seed production.
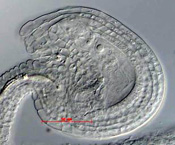
Whole-mount preparation showing an unfertilized mature embryo sac
The alternation of a gametophytic haploid generation with a sporophytic diploid generation is a fundamental aspect of plant
reproduction. In flowering plants, the male gametophytes (the pollen grains) develop within the anthers, and the female
gametophytes (the embryo sacs) develop within the ovule. Fertilization of the egg cell within the embryo sac by one of the
sperm cells delivered by the pollen tube results in the diploid zygote, which initiates the next sporophytic generation.
Although development and function of the gametophytes are critical for plant reproduction, relatively little is known about
the genes required for, and the pathways involved in gametophytic development in flowering plants. In this project, we propose
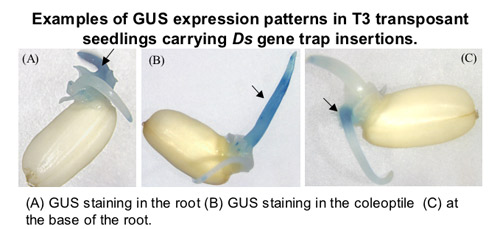
to identify and determine functions of gametophytic genes in Arabidopsis . A mutant screen for gametophytic mutations using Ds
insertion lines has been performed, resulting in the identification of ~ 300 gametophytic mutants. The genes disrupted by these
insertions are being identified and the sequences will be posted on this website. The identification of genes expressed in the
female gametophyte will also be carried out by the hybridization of an Arabidopsis "whole genome" oligoarray using RNA from wild
type and from mutant ovules without embryo sacs. As a result, characterization of gene function is carried out through integrated
genetic, cellular and developmental studies of the female and male gametophytes in the mutants. The characterized mutants will be
made publicly available through deposition in the ABRC stock center.
For information on the results for the lines that have aberrant transmission through the male gametophytes, go to the McCormick lab website.
Identification of gametophytic genes through mutant screens
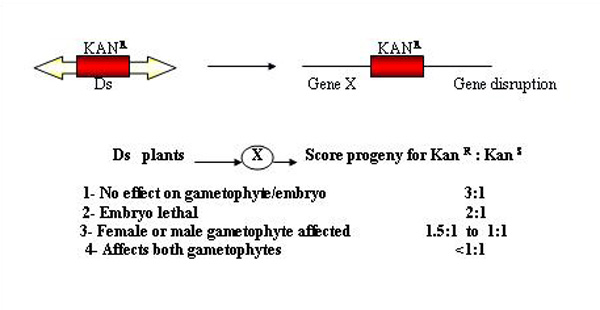
Screening for gametophytic mutations by reduced transmission of kan R in Ds lines
Characterization of female gametophyte defects in the mutant
Megagametogenesis involves the establishment of the functional megaspore, three rounds of mitosis, establishment of female
gametophyte polarity, central vacuole formation, nuclear migration, cellularization, cell specification and differentiation,
nuclear fusion of the two polar nuclei and cell death of the antipodal cells and of the synergid cells. Mutations in the genes
implicated in any of these processes can yield a female gametophyte mutant. In addition, mutants that have no visible defects
in the embryo sacs, but that exhibit problems in pollen tube guidance, fertilization, or early embryo development can be observed.
In order to study the nature of the female defect, whole mount ovules are initially observed by Nomarski/DIC microscopy after
clearing, allowing relatively rapid observations of large numbers of ovules at different stages of development. Preliminary
observations showed that mutants with reduced female transmission were of three general types: (1) Defective in megaspore
specification or function; (2) Defective in embryo sac development and (3) Embryo sac development appears normal, but defective
in subsequent fertilization and/or embryogenesis.

Phenotype of mutants observed by DIC microscopy A, Embryo sac development is arrested at the 2 nuclear stage. Arrowheads indicate the nuclei. B, Embryo sac development is arrested at the 4 nuclear stage. C, Mature wild-type embryo sac. D, Polar nuclei fail to fuse (polar nuclei are indicated with arrows). E , embryo development arrested at 1 cell zygotic stage (arrow). Endosperm development is also affected (endosperm nuclei are indicated by arrowheads). F, Wild-type control embryo at same stage.
Identification of female gametophyte-specific genes using oligonucleotides microarrays
Since gametophyte development occurs within the sporophyte, the isolation of RNA from developing gametophytes without
contamination with the maternal tissues represents a technical challenge. We previously described a gene called SPOROCYTELESS
(SPL) required for initiation of gametophyte development in Arabidopsis (Yang et al. 1999). Plants that are homozygous for the
spl mutation have ovules that appear normal except that they do not contain embryo sacs, and the nucellus does not degenerate.
Therefore, the spl mutant provides an opportunity to identify female gametophyte-specific genes by expression profiling.
Oligonucleotide microarrays are being used to identify genes specific for the female gametophyte by comparison of the expression
profiles of wild-type ovules (embryo-sac + ) with spl mutant ovules (embryo sac - ).
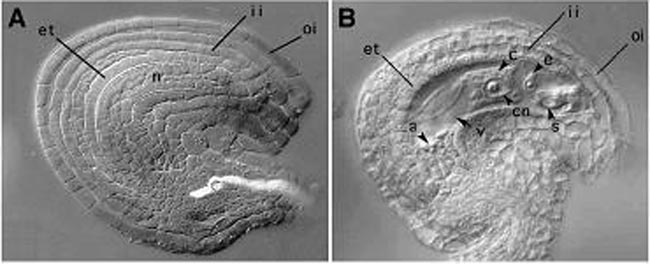
Embryo sac development is arrested at the 2 nuclear stage. Arrowheads indicate the nuclei. B, Embryo sac development is arrested at the 4 nuclear stage. C, Mature wild-type embryo sac. D, Polar nuclei fail to fuse (polar nuclei are indicated with arrows). E , embryo development arrested at 1 cell zygotic stage (arrow). Endosperm development is also affected (endosperm nuclei are indicated by arrowheads). F, Wild-type control embryo at same stage.
Publications